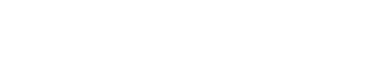Research
We are developing novel technologies to analyze important problems in biology. Details of those technologies are as follows. We are also developing other novel technologies eagerly.
1. Locus-specific chromatin immunoprecipitation (locus-specific ChIP) technologies consisting of iChIP and enChIP
Comprehensive understanding of mechanisms of genome functions, such as epigenetic regulation and transcription, requires the identification of molecules that bind to the genomic regions of interest in vivo. However, non-biased methods to identify the molecules bound to specific genomic loci in vivo are limited. To perform biochemical and molecular biological analyses of specific genomic regions, we developed locus-specific chromatin immunoprecipitation (locus-specific ChIP) technologies that comprise insertional ChIP (iChIP) and engineered DNA-binding molecule-mediated ChIP (enChiP) to purify the genomic regions of interest. After purification of the target genomic region while retaining the molecular interactions, interacting proteins can be identified using mass spectrometry (MS), whereas interacting RNAs and other genomic regions can be identified through next-generation sequencing (NGS).
The scheme of iChIP is as follows: (i) A repeat of the recognition sequence of an exogenous DNA-binding protein, such as LexA, is inserted into the genomic region of interest in the cells to be analyzed. (ii) The DNA-binding domain (DNA DB) of the exogenous DNA-binding protein is fused with a tag(s) and a nuclear localization signal (NLS)(s), and expressed into the cells to be analyzed. (iii) The resultant cells are stimulated and crosslinked with formaldehyde or other crosslinkers, if necessary. (iv) The cells are lysed, and the crosslinked DNA is fragmented either by using sonication or digested with nucleases, such as restriction enzymes. (v) The complexes including the exogenous DNA DB are immunoprecipitated using an antibody against the tag. (vi) The isolated complexes retain molecules interacting with the genomic region of interest. Reverse crosslinking and subsequent purification of DNA, RNA, proteins, or other molecules allows their identification and characterization (Fig. 1).
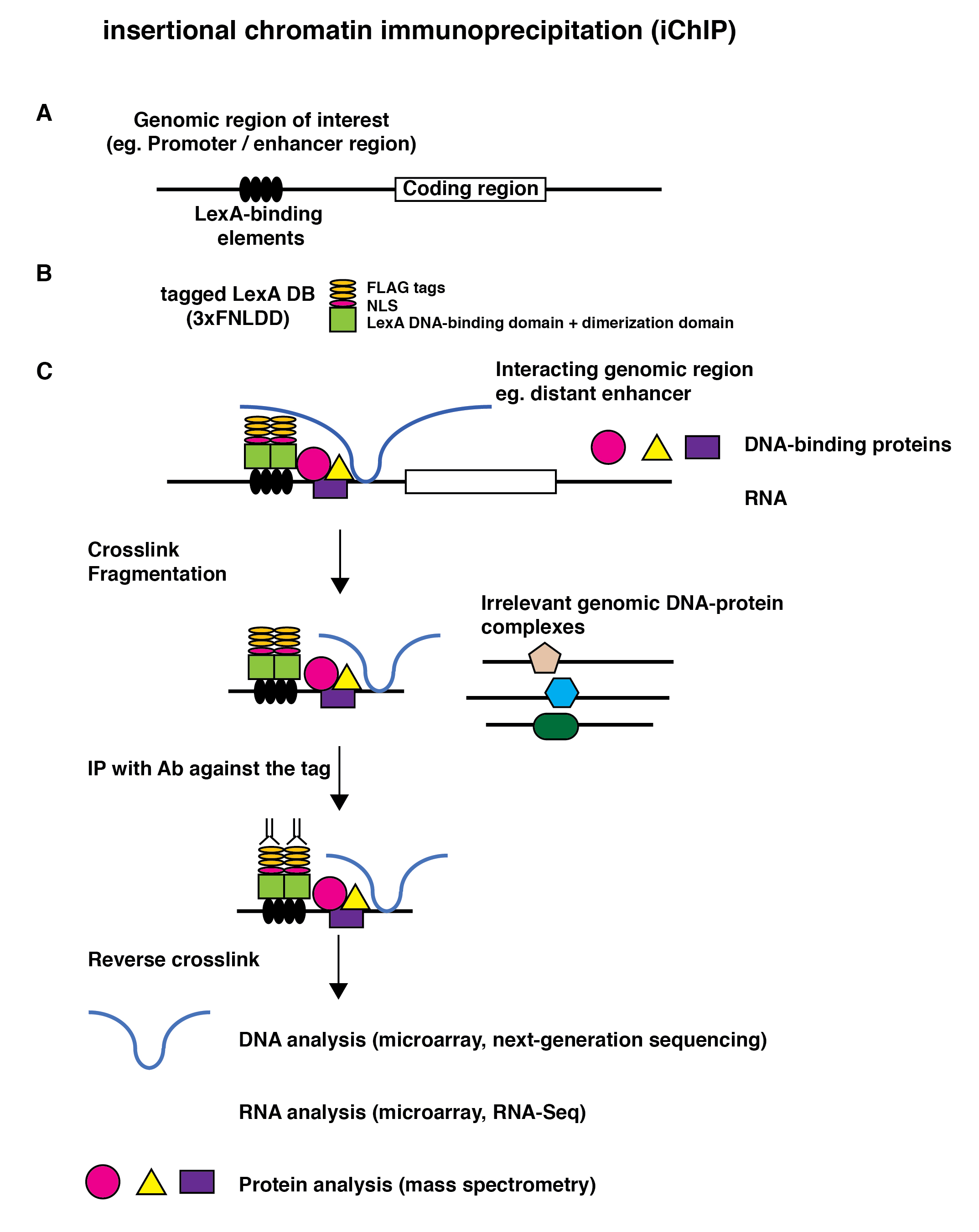
The scheme of enChIP is as follows: (i) A DNA-binding molecule / complex (DB) is generated to recognize DNA sequence in the genomic region of interest. This can be achieved by utilizing technologies, such as engineered zinc-finger proteins, transcription activator-like (TAL) proteins, and the CRISPR system consisting of dCas9 together with gRNA. The engineered DB is fused with a tag(s) and an NLS(s), and expressed in the cells to be analyzed. (ii) The resultant cells are stimulated and crosslinked with formaldehyde or other crosslinkers, if necessary. (iii) The cells are lysed, and DNA is fragmented. (iv) The complexes including the engineered DB are subjected to affinity purification, such as immunoprecipitation. (v) The isolated complexes retain molecules interacting with the genomic region of interest. Reverse crosslinking and subsequent purification of DNA, RNA, proteins, or other molecules allows the identification and characterization of these molecules (Fig. 2). Unlike iChIP, enChIP does not require insertion of recognition sequences of exogenous DNA-binding proteins, making the procedure much more straightforward.
Additionally, we developed in vitro iChIP and in vitro enChIP. These methods utilize recombinant exogenous DNA-binding proteins, such as LexA / dCas9 and synthetic gRNA to tag the target genomic regions in vitro and do not require steps of expressing exogenous DNA-binding molecules. Especially, in vitro enChIP enables the application of locus-specific ChIP to the samples, such as clinical samples and pathogenic microbes, in which the expression of exogenous molecules is difficult.The locus-specific ChIP technology enables non-biased identification of the molecules that bind to even a single copy of target genomic regions, and thus, can be implemented for the identification of novel drug targets.
2. ORNi-PCR and related technologies
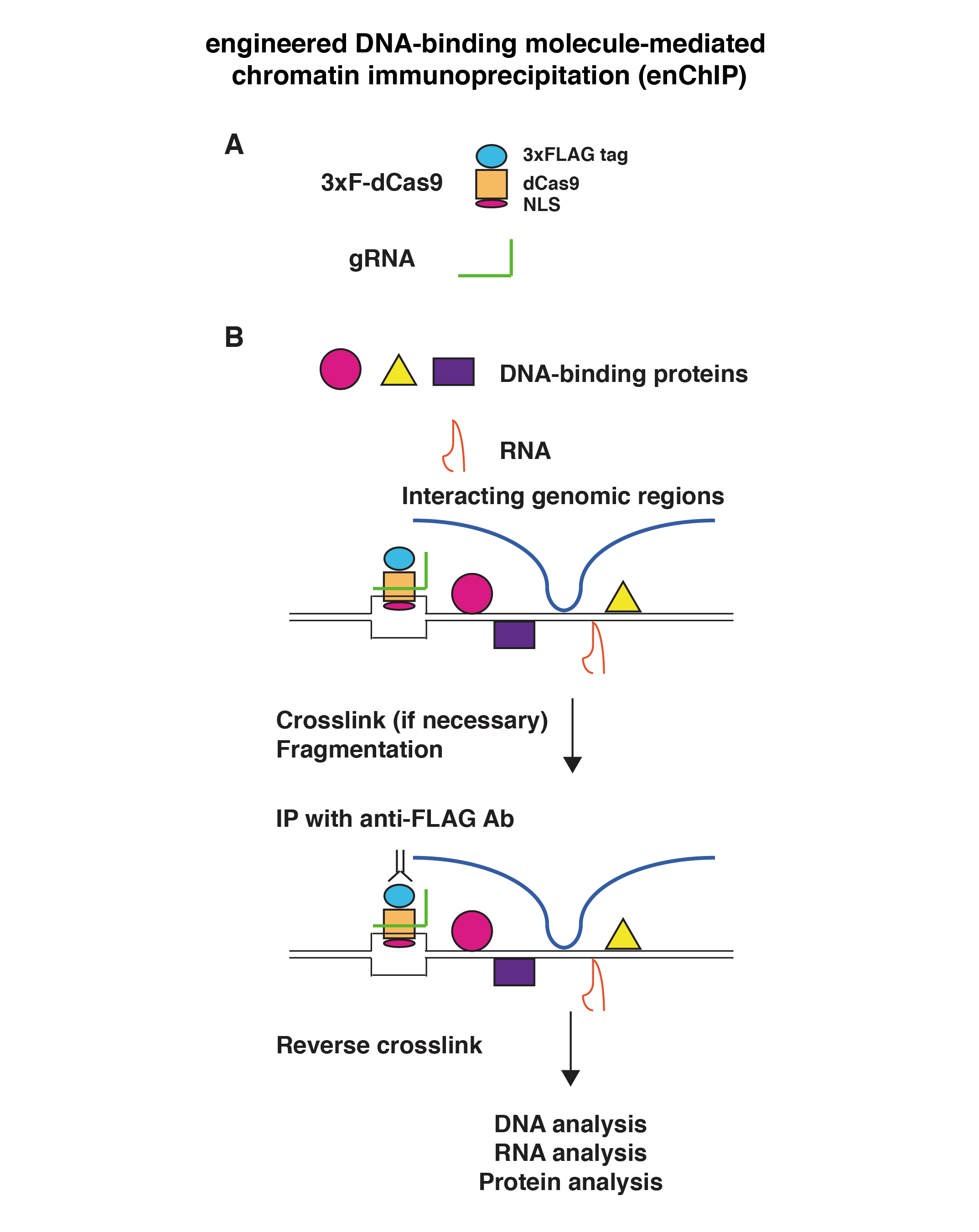
DNA sequence variants are known to be involved in the emergence of intractable diseases, such as cancer. Therefore, various methods using PCR, a method for amplifying DNA, have been employed to detect the presence or absence of DNA mutations known to cause these diseases. However, conventional PCR-based methods have several limitations, such as those regarding detection sensitivity, cost, and ability to provide a definitive diagnosis. We developed a technology called oligoribonucleotide interference-PCR (ORNi-PCR) to overcome these limitations. The ORNi-PCR technology is a method to use an ORN (small RNA), an approximately 17-29 base RNA, to specifically inhibit the amplification of a DNA domain complementary and thus hybridized to the ORN in order to halt a PCR (Fig. 3). Conversely, ORNi-PCR enables the specific amplification of DNA with mutations in domains hybridized with an ORN. ORNi-PCR does not always require information regarding mutation sequences if the target DNA sequence and mutation site to be suppressed for amplification are defined in advance. ORNi-PCR has been reported to be able to distinguish single base mutations with high precision and sensitivity and has thus been used for applications, such as detecting genome-edited cells, DNA methylation analysis, and bacterial flora analysis.